Anat_mriqc pipeline¶
Get no-reference IQMs (image quality metrics) from structural (T1w and T2w) data using mriqc anatomical workflow (mriqc v22.06)¶
Disclaimer: A special effort has been made to provide a pipeline in Mia that gives as much as possible the same result as when computing with the native MRIQC. The variation in results between multiple runs of the same inputs is the result of random sampling and floating point precision errors. These variations are usually small, but if an identical result is sought between multiple runs, the environment variable ANTS_RANDOM_SEED should be set (e.g. ANTS_RANDOM_SEED = 1). If you want obtain the same results as with MIA by using a “Bare-metal” installation of the native MRIQC on your computer, the environment variables ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS and OMP_NUM_THREADS should be set (e.g. ITK_GLOBAL_DEFAULT_NUMBER_OF_THREADS = 1 and OMP_NUM_THREADS = 1). Further discussion is available in a mia_processes ticket.
Pipeline insight
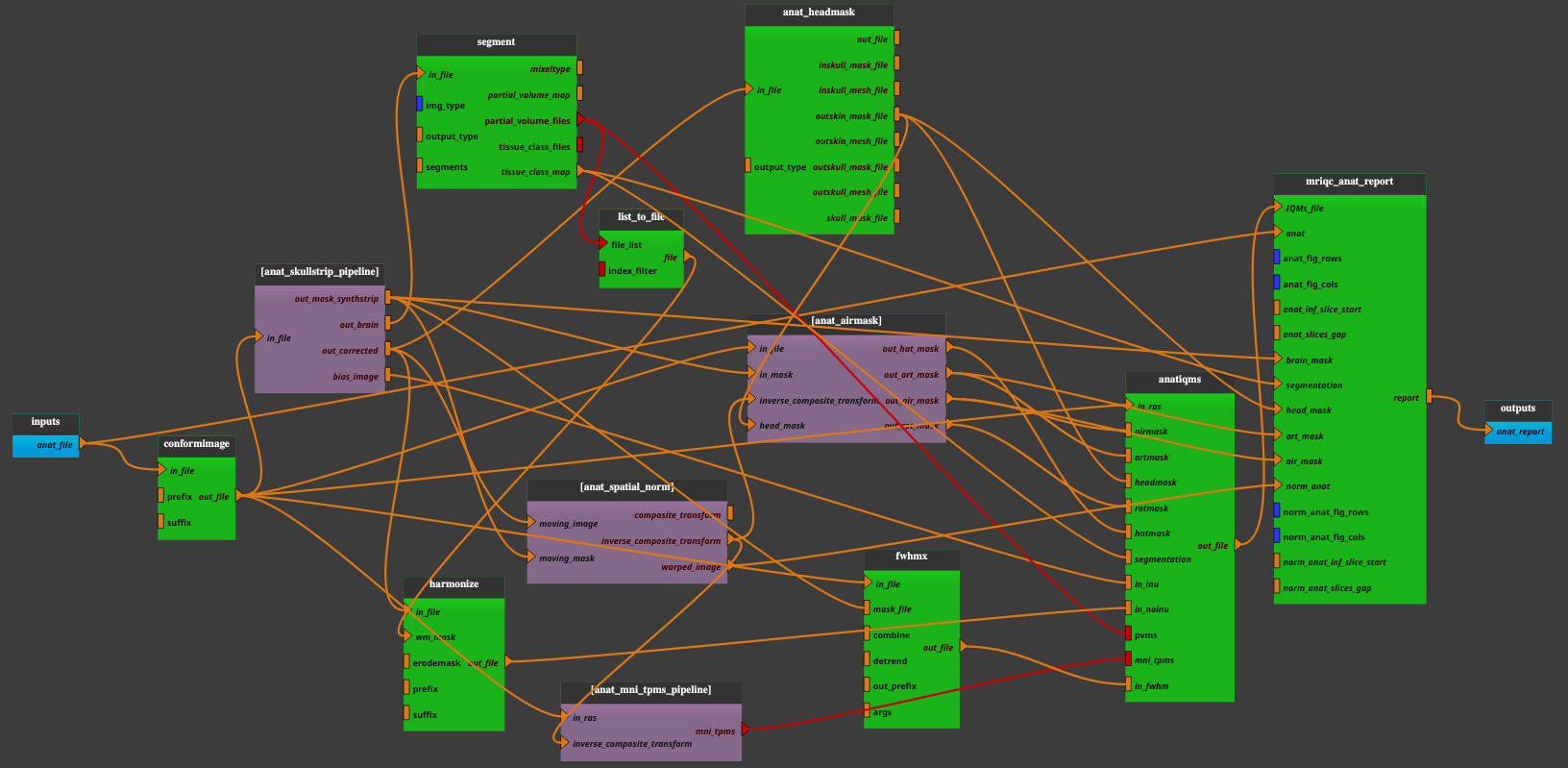
Mandatory inputs parameters
- anat_file (a string representing an existing file)
An anatomical image (T1w or T2w). An existing file (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w.nii'
Outputs parameters:
- anat_report
Anatomical Image-Quality Metrics summary report.
ex. '/home/username/data/derived_data/T1w_ref_anatomical_mriqcReport_2023_03_31_11_42_10_75.pdf'
Useful links: