Anat_airmask pipeline¶
Compute rotation mask, air mask, artifact mask and hat mask for structural data following step 1 from [Mortamet2009]¶
Adapted from mriqc anatomical workflow.
Pipeline insight
Anat_airmask pipeline combines the following pipelines and processes:
(default values: interpolation = ‘MultiLabel’)
(default values : in_template = ‘MNI152NLin2009cAsym’, resolution = 1, suffix = ‘mask’, desc = ‘head’)
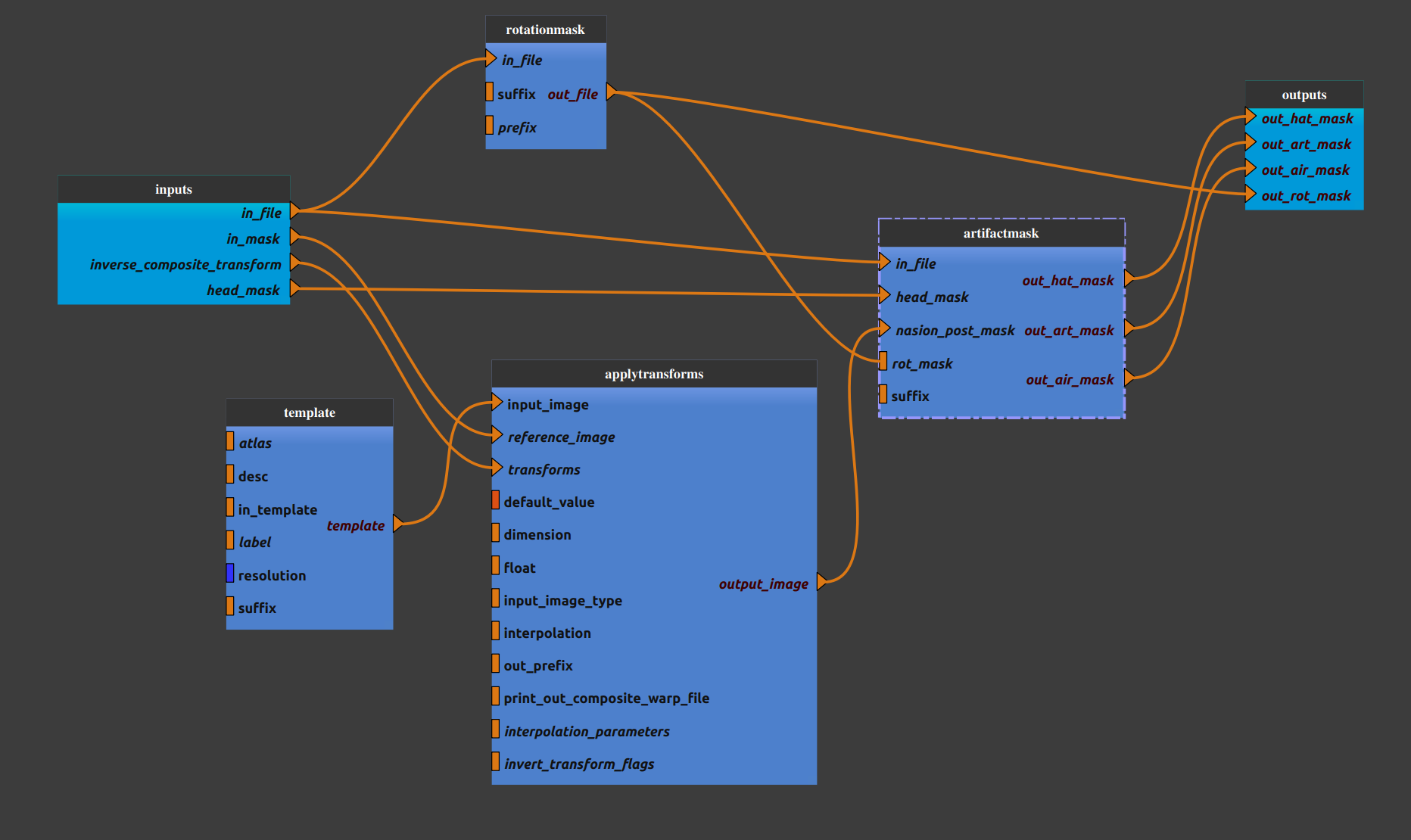
Mandatory inputs parameters
- head_mask (a string representing an existing file)
Head mask (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w_brain_outskin.nii'
- inverse_composite_transform (a string representing an existing file)
Inverse composite transform used in “ApplyTransform” process (MNI space –> in_file space).
ex. '/home/username/data/raw_data/T1w_masked_InverseComposite.h5'
- in_file (a string representing an existing file)
An anatomical image (T1w or T2w) (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w.nii'
- in_mask (a string representing an existing file)
Mask image used as reference image in “ApplyTransform” process (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w_brain_mask.nii'
Outputs parameters:
- out_air_mask
Air mask.
ex. '/home/username/data/derived_data/air_T1w_mask.nii'
- out_art_mask
Artifact mask.
ex. '/home/username/data/derived_data/art_T1w_mask.nii'
- out_hat_mask
Hat mask.
ex. '/home/username/data/derived_data/hat_T1w_mask.nii'
- out_rot_mask
Rotation mask.
ex. '/home/username/data/derived_data/rot_T1w_mask.nii'
Useful links: