Anat_spatial_norm pipeline¶
Spatial normalization to MNI (using MNI152NLin2009cAsym template)¶
Adapted from mriqc 22.06 anatomical workflow
Pipeline insight
Anat_spatial_norm pipeline combines the following pipelines and processes:
(default values for template : in_template = ‘MNI152NLin2009cAsym’, resolution = 2, suffix = ‘T1w’,
default values for template mask : in_template = ‘MNI152NLin2009cAsym’, resolution = 2, suffix = ‘mask’, desc =’brain’)
- Mask
(default values:
convergence_threshold= [1e-07, 1e-08],
convergence_window_size = [15, 5, 3],
interpolation = ‘LanczosWindowedSinc’,
metric = [‘Mattes’, ‘Mattes’],
metric_weight = [1.0, 1.0],
number_of_iterations = [[20], [15]],
radius_or_number_of_bins = [56, 56],
sampling_percentage = [0.2, 0.1],
sampling_strategy = [‘Random’, ‘Random’],
shrink_factors = [[2], [1]],
smoothing_sigmas = [[4.0], [2.0]],
transform_parameters = [(1.0,),(1.0,)],
transforms = [‘Rigid’, ‘Affine’],
use_histogram_matching = [False,True])
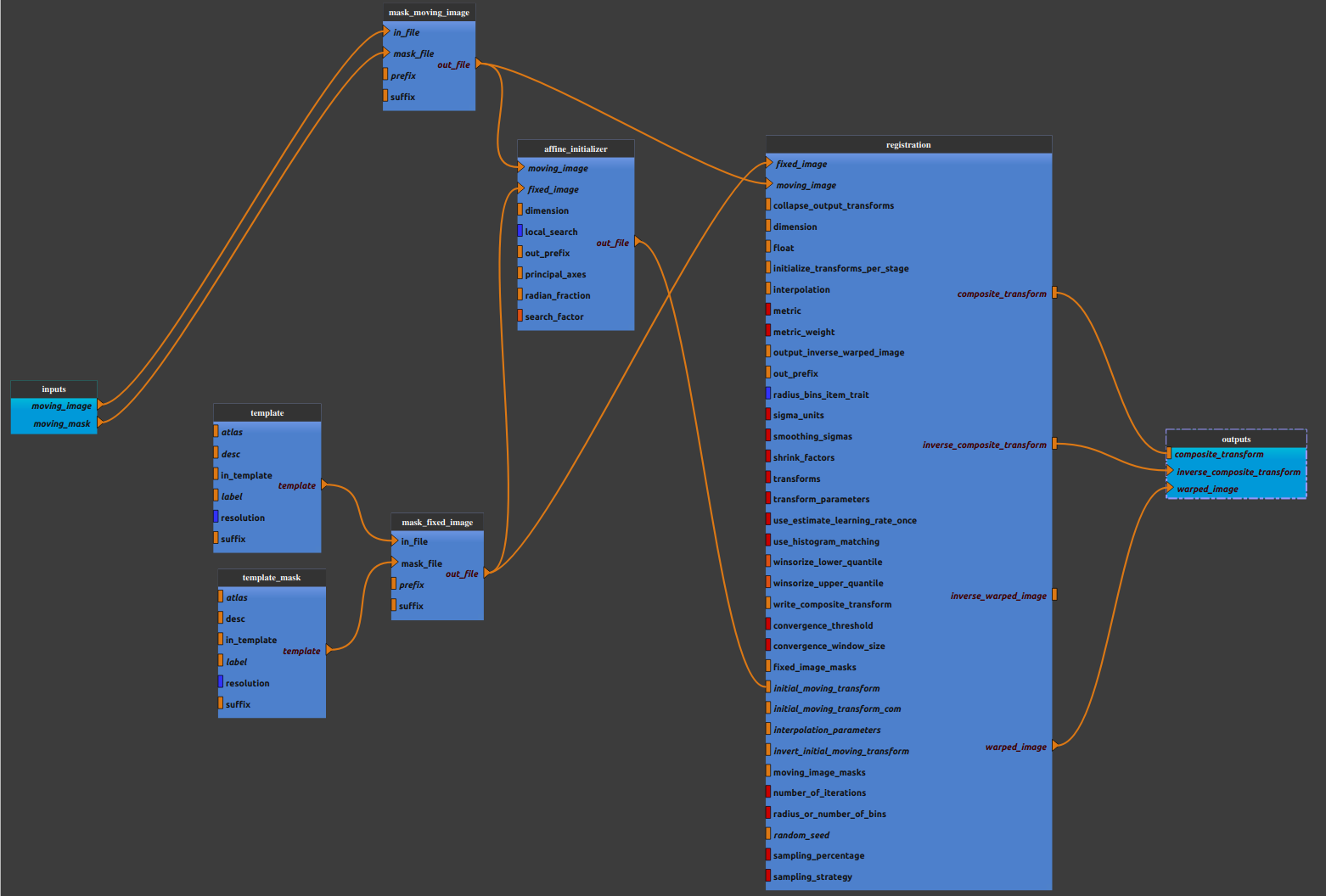
Mandatory inputs parameters
- moving_image (a string representing an existing file)
Anatomical image (T1w or T2w) to register in MNI space (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w.nii'
- moving_mask (a string representing an existing file)
Brain mask used to mask moving image (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w_desc-brain_mask.nii'
Outputs parameters:
- composite_transform
Composite transform (moving_image masked space –> MNI)
ex. '/home/username/data/derived_data/T1w_masked_Composite.h5'
- inverse_composite_transform
Inverse composite transform (MNI –> moving_image masked space)
ex. '/home/username/data/derived_data/T1w_masked_InverseComposite.h5'
- warped_image
Masked moving image in template space.
ex. '/home/username/data/derived_data/w_T1w_masked.nii'
Useful links: