Bold_stat_cvr brick¶
SPM-based first level GLM analysis used in the CVR evaluation¶
Pipeline insight
- Bold_stat_cvr pipeline combines the following bricks:
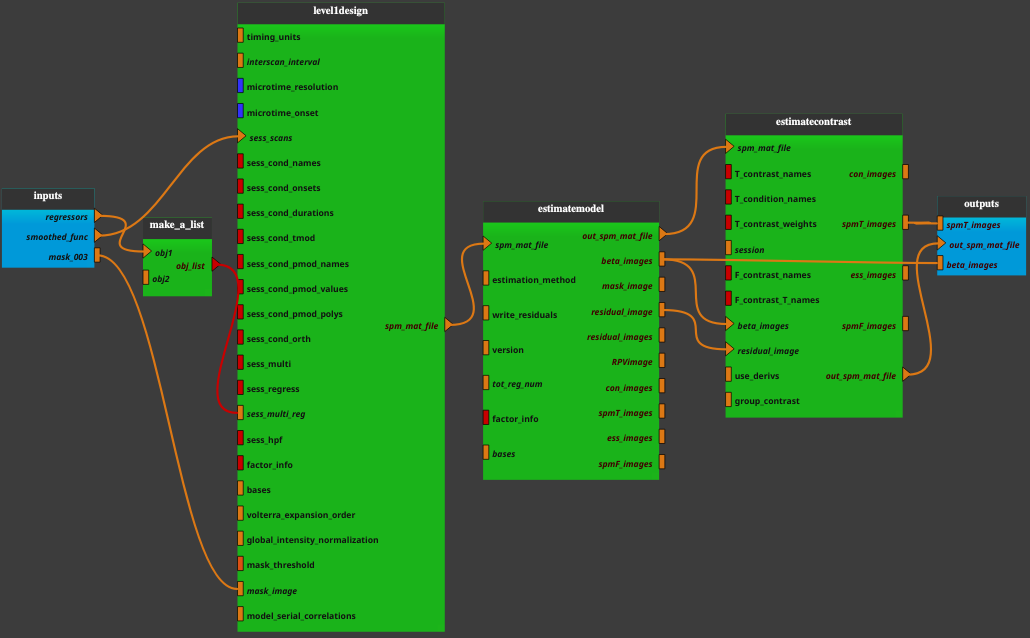
Inputs parameters:
- regressors
A list of *.mat/*.txt file(s) containing details of multiple regressors. *.mat files must contain a matrix R and *.txt file contain directly a regressor (see SPM manual for further explanation [1]).
ex. ['/home/username/data/raw_data/rp_func.txt', '/home/username/data/raw_data/regressor_physio_EtCO2_ctl.mat']
- smoothed_func
A previously preprocessed functional images (a list of items which are a pathlike object or string representing an existing file).
ex. ['/home/username/data/raw_data/swrfunc.nii']
- mask_003
A grey matter mask image with a resolution defined previously in the Spatial_mask.
ex. /home/username/data/raw_data/mask_anat_002.nii
Outputs parameters:
- spmT_images
Statistic image from a t-contrast (t-statistic maps).
ex. /home/username/data/derived_data/spmT_0001.nii
- out_spm_mat_file
The final SPM.mat file containing all the details of the analysis.
ex. /home/username/data/derived_data/SPM.mat
- beta_images
A list of images. One image is created for each column in the design matrix, and the value at each voxel within these images represents the beta value for that predictor at that voxel.
ex. ['/home/username/data/derived_data/beta_0001.nii', '/home/username/data/derived_data/beta_0002.nii', '/home/username/data/derived_data/beta_0003.nii', '/home/username/data/derived_data/beta_0004.nii', '/home/username/data/derived_data/beta_0005.nii', '/home/username/data/derived_data/beta_0006.nii', '/home/username/data/derived_data/beta_0007.nii', '/home/username/data/derived_data/beta_0008.nii']