Spatial_mask pipeline¶
From a list of images resulting from a previous segmentation, produces a grey matter mask¶
Pipeline insight
- Spatial_mask pipeline combines the following bricks:
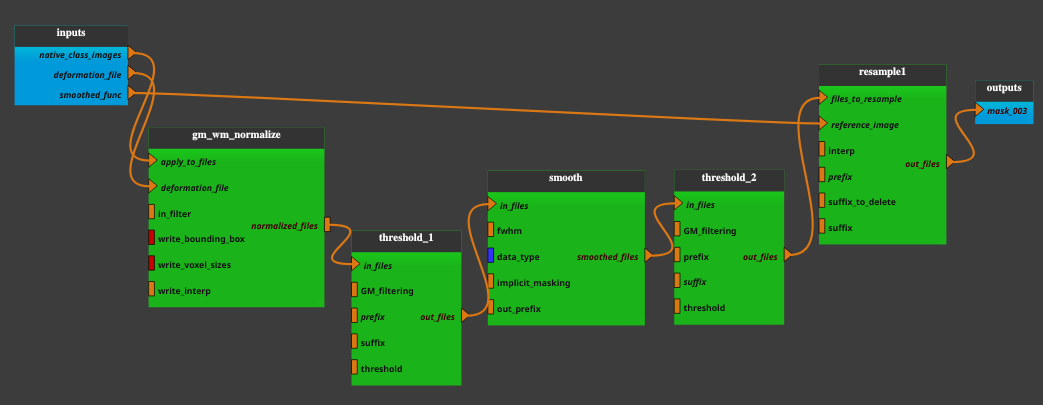
inputs parameters:
- native_class_images
A list of images, resulting from a previous segmentation to grey matters, white matters, etc. (valid extensions: [.nii, .nii.gz]).
ex. ['/home/username/data/raw_data/c1anat.nii', '/home/username/data/raw_data/c2anat.nii', '/home/username/data/raw_data/c3anat.nii', '/home/username/data/raw_data/c4anat.nii', '/home/username/data/raw_data/c5anat.nii', '/home/username/data/raw_data/c6anat.nii']
- deformation_file
File y_*.nii containing 3 deformation fields for the deformation in x, y and z dimension (a pathlike object or string representing an existing file (valid extensions in [.img, .nii, .hdr]).
ex. /home/username/data/raw_data/y_anat.nii
- smoothed_func
A functional image (valid extensions: [.nii, .nii.gz]).
ex. /home/username/data/raw_data/func.nii
Outputs parameters:
- mask_003
The grey matter mask at the resolution of the smoothed_func.
ex. /home/username/data/derived_data/mask_swc1anat_003.nii
- NOTE:
The selection of the images is made using the syntax of SPM (c1* for grey matter and c2* for white matter).