Dwi_tissue_boundaries pipeline¶
Create tissue boundaries with a T1w and registratiojn in DWI space¶
The aim of this pipeline is to prepare Anatomically Constrained Tractography (ACT) to increase the biological plausibility of streamline creation when tracking will be performed.
Note that this pipeline used the FLIRT registration (FSL) to co-register diffusion and anatomical data as suggested in the B.A.T.M.A.N. tutorial. However it seems to not work perfectly for some data. Please check the outputs data.
Pipeline insight
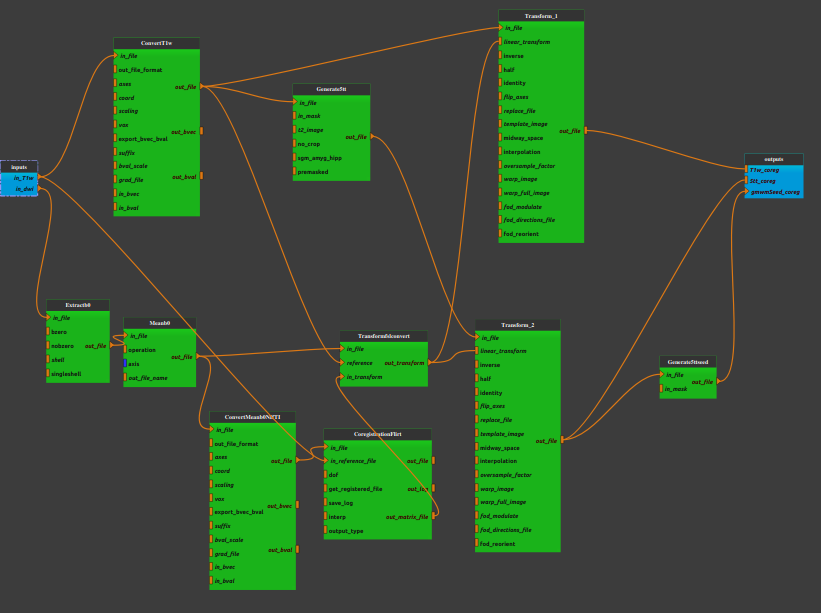
Mandatory inputs parameters
- in_dwi (a string representing an existing file)
Preprocessed diffusion image (valid extensions: [.mif]).
ex. '/home/username/data/raw_data/DWI_denoised_unringed_dwifslpreproc_unbias.nii'
- in_T1w (a string representing an existing file)
Anatomical image (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/T1w.nii'
Outputs parameters:
- T1w_coreg
Anatomical image coregistered in DWI space.
ex. '/home/username/data/derived_data/T1w_transformed.mif'
- 5tt_coreg
5tt image (a 4-dimensional preprocessed T1-weighted image with 5 different tissue types (cortical gray matter, subcortical gray matter, white matter, CSF and pathological tissue)) coregistered in DWI space.
ex. '/home/username/data/derived_data/T1w_5tt_transformed.mif'
- gmwmSeed_coreg
A mask of the gray-matter/white-matter-boundary in DWI space.
ex. '/home/username/data/derived_data/T1w_5tt_transformed_gmwmSeed.mif'