Dwi_whole_brain_tractograpy pipeline¶
Brain Tractography with MRTrix¶
The aim of this pipeline is to create a whole-brain tractogram for a multishell DWI using Constrained Spherical Deconvolution (CSD). In this pipeline, an image with b=0 volumes with opposite phase encoding should be provided for the purpose of EPI distortion correction. An anatomical image is also required.
The pipeline is based on B.A.T.M.A.N. tutorial and Andy’s brain book tutorial
Note that this pipeline used the FLIRT registration (FSL) to co-register diffusion and anatomical data as suggested in the B.A.T.M.A.N. tutorial. However it seems to not work perfectly for some data.
Test this pipeline : use the sub-002 from mia_data_users.
Pipeline insight
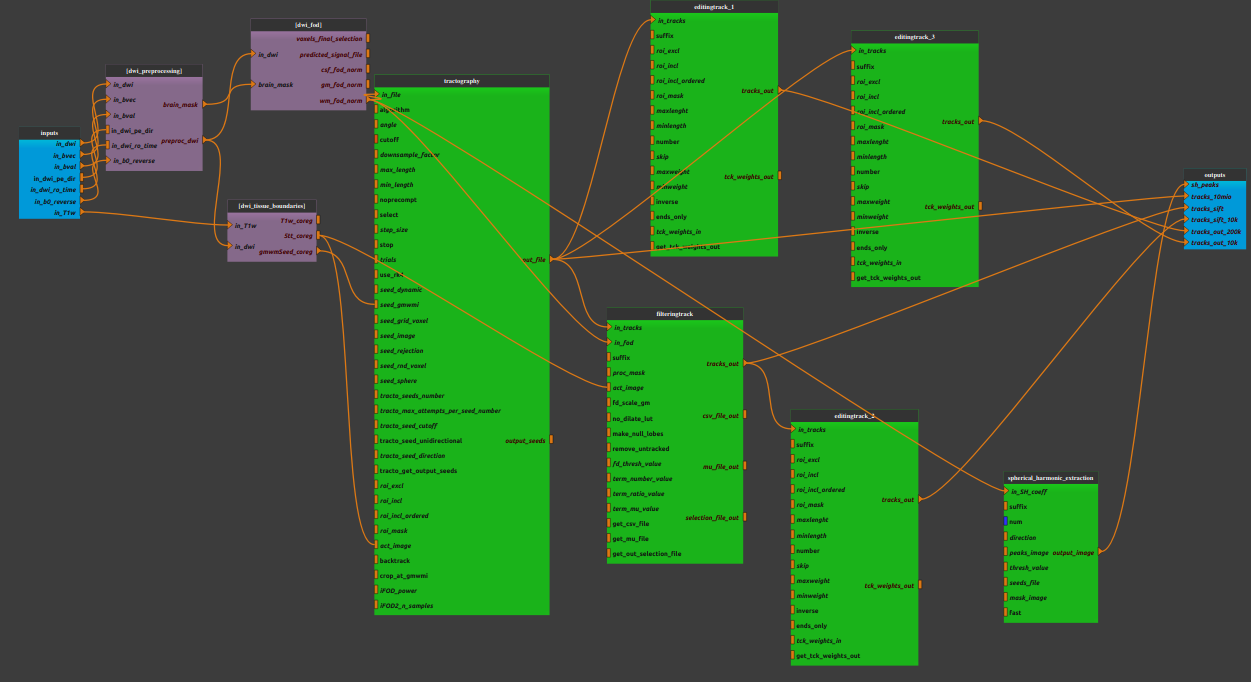
Mandatory inputs parameters
- in_dwi (a string representing an existing file)
Diffusion image to preprocess (valid extensions: [.nii, .nii.gz, .mif]). If a NIfTI is supplied, bvec and bval files will be found automatically.
ex. '/home/username/data/raw_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000.nii'
- in_dwi_pe_dir (ap, pa, lr, rl, default value is ap, optional)
- Phase encoding direction of the in_diw image:- ap : Anterior to posterior- pa: Posterior to anterior- lr: Left to right- rl: Right toleft
ex. pa
- in_dwi_ro_time (a float, optional)
Total readout time of in_dwi image (in seconds).
ex. 0.0298634
- in_b0_reverse (a string representing an existing file)
b=0 volumes with opposing phase-direction which is to be used exclusively by topup for estimating the inhomogeneity field (valid extensions: [.nii, .nii.gz, .mif]).
ex. '/home/username/data/raw_data/sub-002-ses-01--1-DWI-APA_0_Axi2_50_EnhG-DwiSE-151925_410000.nii'
- in_T1w (a string representing an existing file)
Anatomical image (valid extensions: [.nii, .nii.gz]).
ex. '/home/username/data/raw_data/sub-002-ses-01--2-T1_3D-T1TFE-150551_470000.nii'
Outputs parameters:
- sh_peaks (a string representing a file)
The peaks of a spherical harmonic function in each voxel Each volume corresponds to the x, y & z component of each peak direction vector in turn.
ex. '/home/username/data/derived_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000_denoised_unringed_dwifslpreproc_unbias_wm_odf_norm_peaks.mif'
- tracks_10mio (a string representing a file)
The tractography with 10 million streamlines
ex. '/home/username/data/derived_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000_denoised_unringed_dwifslpreproc_unbias_wm_odf_norm_tracto.tck'
- tracks_200k (a string representing a file)
The reduce tractography with 200k streamlines
ex. '/home/username/data/derived_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000_denoised_unringed_dwifslpreproc_unbias_wm_odf_norm_tracto_200k.tck'
- tracks_10k (a string representing a file)
The reduce tractography with 10k streamlines
ex. '/home/username/data/derived_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000_denoised_unringed_dwifslpreproc_unbias_wm_odf_norm_tracto_10k.tck'
- tracks_sift (a string representing a file)
The tractography filtered such that the streamline densities match the FOD lobe integrals
ex. '/home/username/data/derived_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000_denoised_unringed_dwifslpreproc_unbias_wm_odf_norm_tracto_sift.tck'
- tracks_sift_10k (a string representing a file)
The reduce sift tractography with 10k streamlines
ex. '/home/username/data/derived_data/sub-002-ses-01--0-DWI-APP_8x0-32x1000-64x2500_Axi2_50_EnhG-DwiSE-151142_650000_denoised_unringed_dwifslpreproc_unbias_wm_odf_norm_tracto_10k.tck'
- T1w_coreg (a string representing a file)
Anatomical image coregistered in DWI space.
ex. '/home/username/data/derived_data/sub-002-ses-01--2-T1_3D-T1TFE-150551_470000_transformed.mif'
Useful links: MRTrix Tutorial B.A.T.M.A.N.: Basic and Advanced Tractography with MRtrix for All Neurophiles Tutorial Andy’s brain book