Remote computing with Popluse-MIA¶
Remote computing is performed using Soma-Workflow.
It needs several install steps before being able to process:
Please note that there are few Limitations.
In these steps, things will be a bit different if the server has to run jobs inside a container such as Singularity. Actually, the computing resource will then need to run the container in jobs, and often to pass it some additional information. The configuration will need to describe this indirection.
There are 3 components working together, all with 2 situations: “native” or “containerized”:
flowchart LR
Client(Client)<-->SWF(Soma-Workflow server)
subgraph client [local client]
Client
end
subgraph server [remote computing resource]
SWF-->Jobs
end
the client (Populse-MIA, Capsul, Axon, or Soma-Workflow GUI) may run natively on the client system, or inside a container.
flowchart LR
Client(Client)<-->SWF(Soma-Workflow server)
subgraph client [local client]
subgraph container
Client
end
end
subgraph server [remote computing resource]
SWF-->Jobs
end
the computing resource should be running Soma-Workflow, either natively, or inside a container - and this is not always possible.
flowchart LR
Client(Client)<-->SWF(Soma-Workflow server)
subgraph client [local client]
Client
end
subgraph server [remote computing resource]
subgraph container
SWF
end
SWF-->Jobs
end
processing jobs may run natively on the computing resource nodes, or inside a container.
flowchart LR
Client(Client)<-->SWF(Soma-Workflow server)
subgraph client [local client]
Client
end
subgraph server [remote computing resource]
SWF-->Jobs
subgraph container
Jobs
end
end
The first point, client running natively or in a container, is normally not a problem and should make no difference. So the discussion will mainly focus on the two last points.
Install Soma-Workflow on the computing server¶
read Soma-Workflow remote execution
Native installation¶
If the computing resource is a cluster with a jobs resource manager (DRMS) like PBS, Grid Engine, Slurm or another one, then this manager needs to be operated by Soma-Workflow, and thus cannot run inside a container: in this situation, Soma-Workflow should be installed natively on the cluster front-end machine.
Python (python3) should be installed and in the PATH of the system
Soma-Workflow can be installed as sources just by cloning the github repository
Ex - On the remote computing login node:
cd
git clone https://github.com/populse/soma-workflow.git
export PYTHONPATH="$HOME/soma-workflow/python:$PYTHONPATH"
then add in your $HOME/.bashrc file:
export PYTHONPATH="$HOME/soma-workflow/python:$PYTHONPATH"
Container installation¶
The client just needs to know how to run it: the client config should specify the PYTHON_COMMAND option. See the client configuration below.
Configure Soma-Workflow on the computing resource¶
See Soma-Workflow documentation.
The configuration file $HOME/.soma-workflow.cfg has to be created or edited on the computing resource side, on the user account. It needs to declare the computing resource, with an identifier.
If the computing server will run jobs inside a container, then each command has to be prefixed with the container run command, such as singularity run /home/myself/container.sif, or /home/myself/brainvisa/bin/bv for a Casa-Distro container like a BrainVisa distribution. This is done using the CONTAINER_COMMAND option, as explained in this documentation.
Ex - On the remote computing login node:
[dr144257@alambic-py3]
SERVER_NAME = dr144257@alambic-py3
# optional limitation of the jobs in various queues
MAX_JOB_IN_QUEUE = {15} test{50} Nspin_long{15}
MAX_JOB_RUNNING = {100} test{500} Nspin_long{50}
container_command = ['/home/dr144257/brainvisa-cea-master/bin/bv']
scheduler_type = pbspro
# native_specification = -l walltime=96:00:00
Configure Soma-Workflow on the client machine¶
See Soma-Workflow documentation.
The configuration file $HOME/.soma-workflow.cfg has to be created or edited on the client system machine side, on the user account. It needs to declare the remote computing resource, with the same identifier it has been declared on the server side.
If the client runs inside a container (such as a Casa-Distro container) using a separate user home directory, then the config file must be located (or symlinked, or mounted) in the container home directory.
If the Soma-Workflow server on the remote computing side should run inside a container, then the client needs to know how to run the container. This is done by specifying the PYTHON_COMMAND option as explained in the remote execution doc. Typically we will use something like:
PYTHON_COMMAND = bv python
or:
PYTHON_COMMAND = /home/myself/brainvisa/bin/bv python
or:
PYTHON_COMMAND = singularity run /home/myself/container.sif python
Ex - On the client local machine, possibly in the container home directory:
[dr144257@alambic-py3]
cluster_address = alambic.intra.cea.fr
submitting_machines = alambic.intra.cea.fr
queues = Default Nspin_run32 Nspin_run16 Nspin_run8 Nspin_run4 Nspin_bigM Nspin_short Nspin_long Cati_run32 Cati_run16 Cati_run8 Cati_run4 Cati_short Cati_long Global_short Global_long run2 run4 run8 run16 run32 lowprio
login = dr144257
allowed_python_versions = 3
Configure the computing resource in Populse-MIA / Capsul¶
Run Populse-MIA:
python3 -m populse_mia
- Go to the menu
File / MIA preferences - In the preferences, open the
Pipelinetab - in the Pipeline tab, click
Edit CAPSUL config - in the Capsul config, go to the
somaworkflowtab type the computing resource name in the
computing_resourceparameter. In our example, type:dr144257@alambic-py3. Well, this is just to set it as the default resource, it’s not mandatory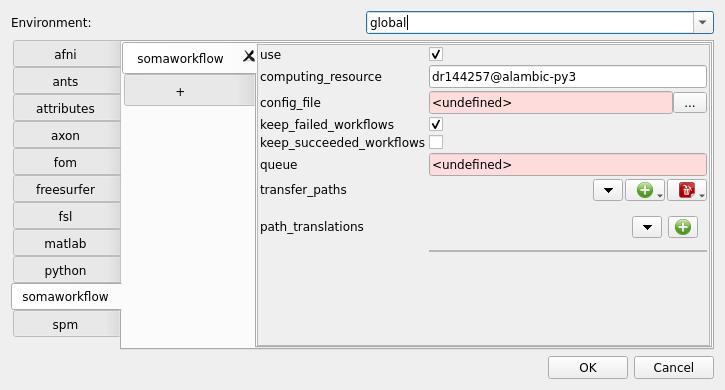
edit the
Environmentparameter at the top of the Capsul config dialog, and enter the computig resource name:dr144257@alambic-py3for us here. Validate by pressing Return: this will create a config entry for this resource.You can enter different config values for this computing resource from the default, “global” one. Especially the
somaworkflowmodule config can be edited to use some data file transfers: some directories can be declared intransfer_paths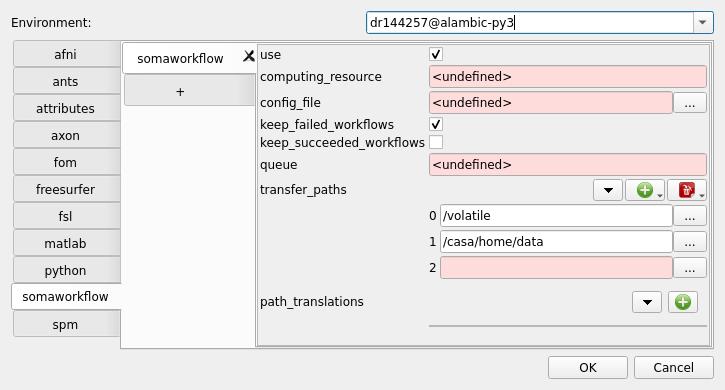
still in the
dr144257@alambic-py3environment (or your computing resource) config, you can set other tabs config, likematlaborspmpaths: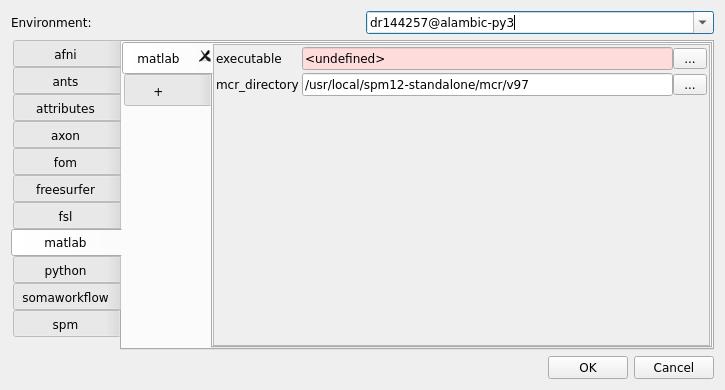
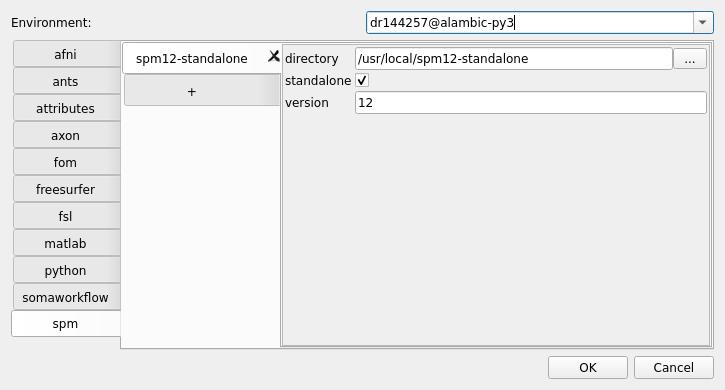
validate by pressing
OKin the Capsul config dialog
- in the Capsul config, go to the
validate the MIA preferences by pressing
OKthere too.
- in the Pipeline tab, click
- In the preferences, open the
- Go to the menu
Run pipelines¶
When Soma-Workflow is enabled, when clicking the Run button in the Pipeline Manager tab of Populse-MIA, then a connection dialog is displayed: it is the classical Soma-Workflow connection dialog:
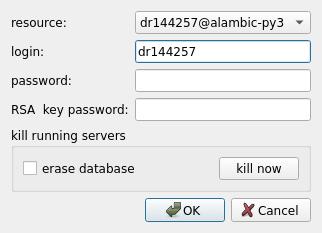
Select the configured resource you want to run the pipeline on, and click OK. The resource will be connected, and the workflow will be sent there. If directories have been declared as transfers, then the input files from these directories will be automatically sent to the remote computing resource (through a secure ssh connection), and results in these directories will be transferred back to the client machine after execution.
You can monitor the execution through the Status button in the Pipeline manager tab - or directly through the soma_workflow_gui monitor program.
In the status window, check the
Soma-Workflow monitoringoption.
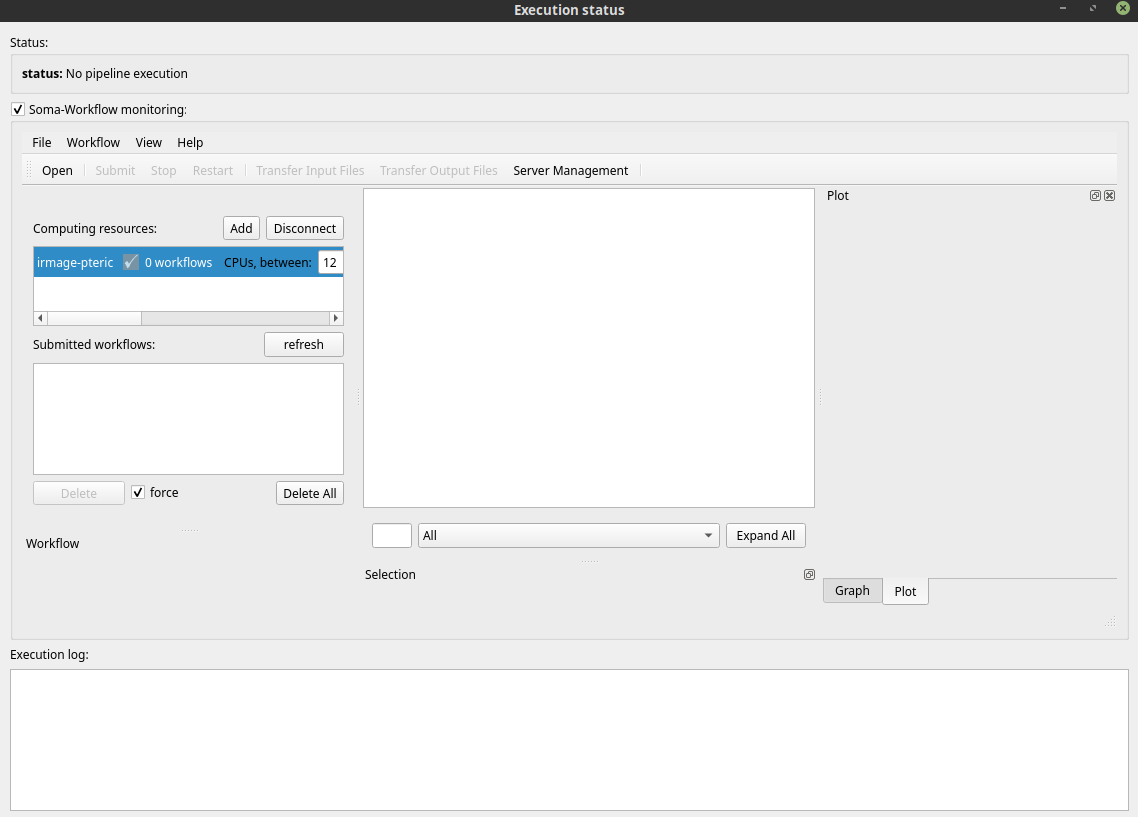
You see… nothing !… Yes it’s normal: you see the local machine and the workflow has been sent to a remote resource: you need to connect the remote monitoring: click the
Addbutton. The same connection dialog appears. Select the resource.After connection, the resource is available. The running workflow should appear first in the list.
Limitations¶
There are a few limitations to the client / server processing
Cluster admins may not like servers running on the login node
Why it’s difficult and often impossible to run the Soma-Workflow server inside a container
Disconnection is partly supported in MIA¶
The pipeline execution engine in MIA is monitoring the execution directly, and when execution is finished, gaterhes the results to index them in the database. If the client is disconnected or shut down before processing has finished, the results indexing will not be done automatically. It will be done partly when clicking the “cleanup” button.
File transfers limitations¶
File transfers are using network bandwidth. For large data, it can be slow enough to forbid any execution.
Transfers are working well as long as they are correctly and fully described in processes input and output parameters. If a process takes a directory as input, the full contents of that directory will be sent to the server. So if the directory contains more than the needed data, it will involve unnecessary, and possibly huge data transfers. In some cases it will simply be impossible to use. An interesting example is SPM / nipype processes which take the output directory as an input parameter: it will likely trigger the transfer of the full database, which is certainly not good.