Bold_spatial_preprocessing3 pipeline¶
An example of fMRI data pre-processing¶
Pipeline insight
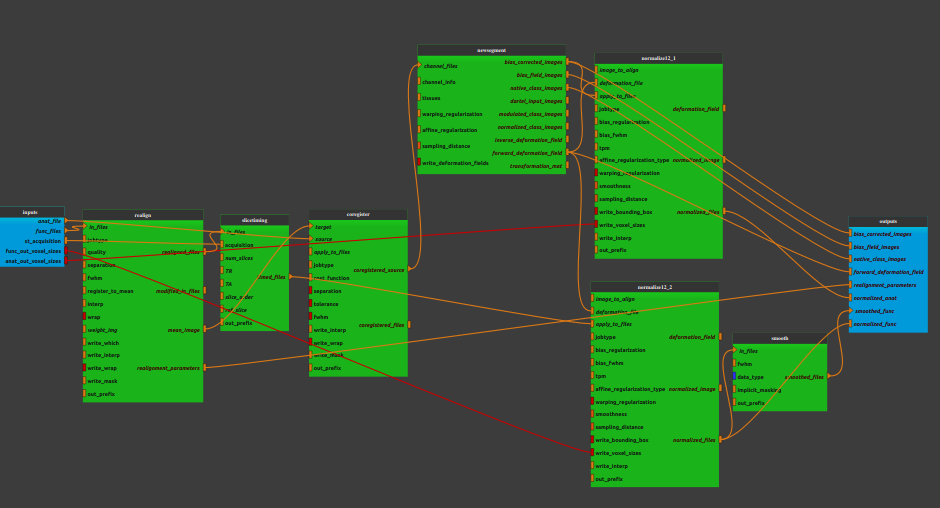
Inputs parameters
- anat_file (an existing uncompressed file):
An anatomical image (valid extensions: [.nii]). Ex. 3D T1 sequence sush as T1 turbo field echo.
ex. '/home/username/data/raw_data/Anat.nii'
- func_files (A list of items which are an existing uncompressed file)
Functional images (valid extensions: [.nii]). Ex. 4D T2* sequence sush as echo planar imaging.
ex. ['/home/username/data/raw_data/Func.nii']
Inputs parameters with default values
- st_acquistion (a string, default is sequential ascending)
Type of the acquisition, either sequential ascending, sequential descending, interleaved (middle-top), interleaved (bottom-up) or interleaved (top-down). Slice ordering is assumed to be from foot to head and bottom slice = 1.
ex. 'sequential ascending'
- func_out_voxel_sizes(a list of three integer, default is [3.0, 3.0, 3.0])
Voxel size of the out functional data (used for write_voxel_size in Normalize12 Write for functional data)
ex. [3.0, 3.0, 3.0]
- anat_out_voxel_sizes(a list of three integer, default is [1.0, 1.0, 1.0])
Voxel size of the out anatomical data (used for write_voxel_size in Normalize12 Write for anatomical data)
ex. [1.0, 1.0, 1.0]
Outputs parameters:
- bias_corrected_images (a list of items which are a pathlike object or string representing an existing file)
The bias corrected images.
ex. '/home/username/data/derived_data/mAnat.nii'
- bias_field_images (a list of items which are a pathlike object or string representing an existing file)
The estimated bias field.
ex. '/home/username/data/derived_data/bias.nii'
- native_class_images (a list of items which are a list of items which are a pathlike object or string representing an existing file)
Native space probability maps .
ex. [['/home/username/data/derived_data/c1Anat.nii'], ['/home/username/data/derived_data/c2Anat.nii'], ['/home/username/data/derived_data/c3Anat.nii'], ['/home/username/data/derived_data/c4Anat.nii'], ['/home/username/data/derived_data/c5Anat.nii']]
- forward_deformation_field (a list of items which are a pathlike object or string representing an existing file)
Forward deformation field. Could be used for spatially normalising images to MNI space.
ex. '/home/username/data/derived_data/y_Anat.nii'
- realignment_parameters (a list of items which are a pathlike object or string representing an existing file)
The estimated translation and rotation parameters during the realign stage.
ex. '/home/username/data/derived_data/rp_Func.txt'
- normalized_anat (a list of items which are a pathlike object or string representing an existing file)
The final normalised anatomical image .
ex. '/home/username/data/derived_data/wAnat.nii'
- normalized_func (a list of items which are an existing file name)
Functional images, realigned, registered with the anatomical image and normalized.
ex. '/home/username/data/derived_data/wrFunc.nii'
- smoothed_func (a list of items which are an existing file name)
The final, realigned then registered with the anatomical image, then normalised then smoothed, functional images .
ex. '/home/username/data/derived_data/swrFunc.nii'